# download most recent software version
#devtools::install_github("geomorphR/geomorph", ref = "Stable", build_vignettes = TRUE)
#devtools::install_github("mlcollyer/RRPP")
# load analysis packages
library(here)here() starts at /Users/seldenjrz/Documents/github/perdiz3/supplibrary(StereoMorph)
library(geomorph)Loading required package: RRPPLoading required package: rglLoading required package: Matrixlibrary(ggplot2)
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(ggpubr)
library(wesanderson)
# read shape data and define number of sLMs
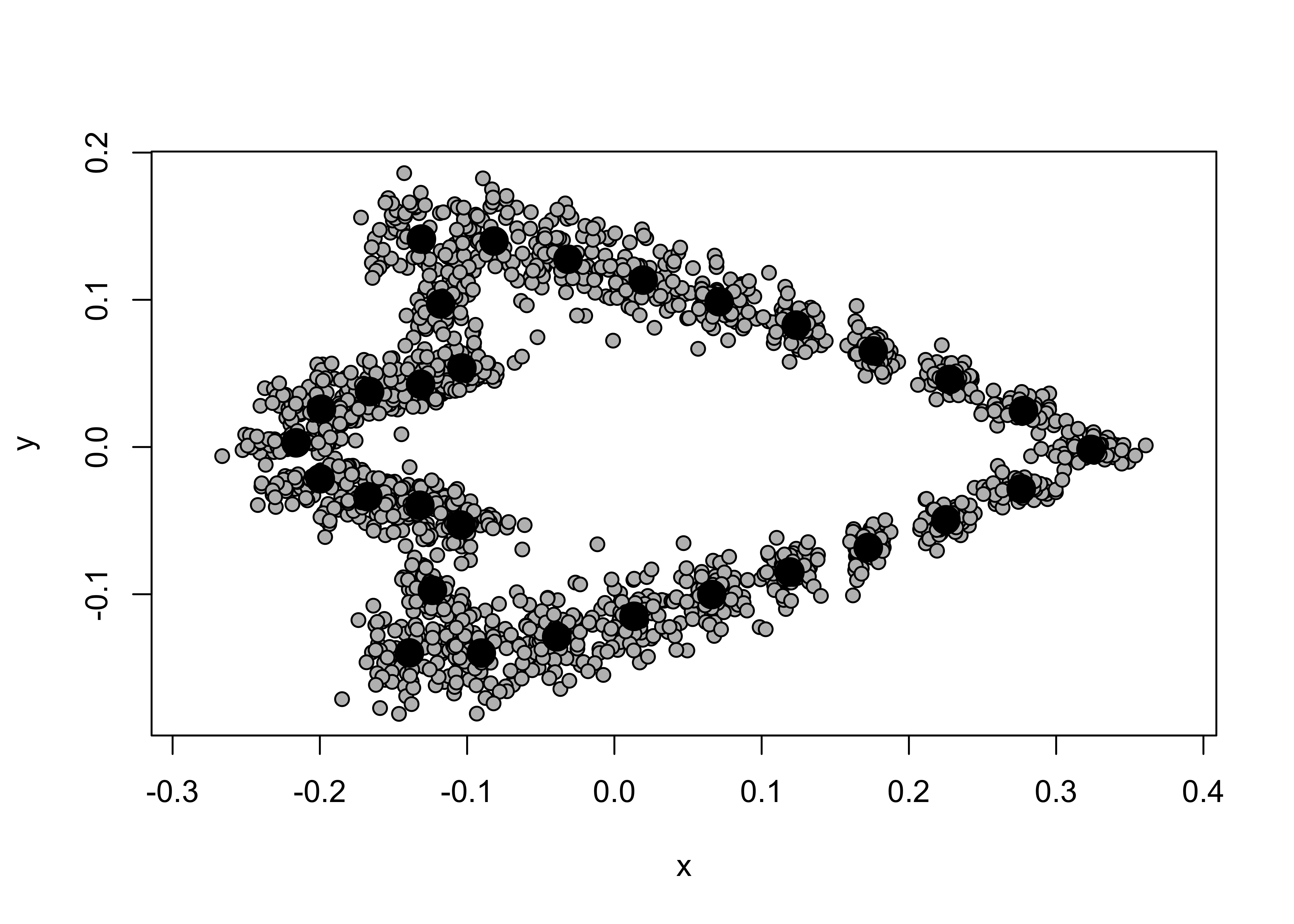
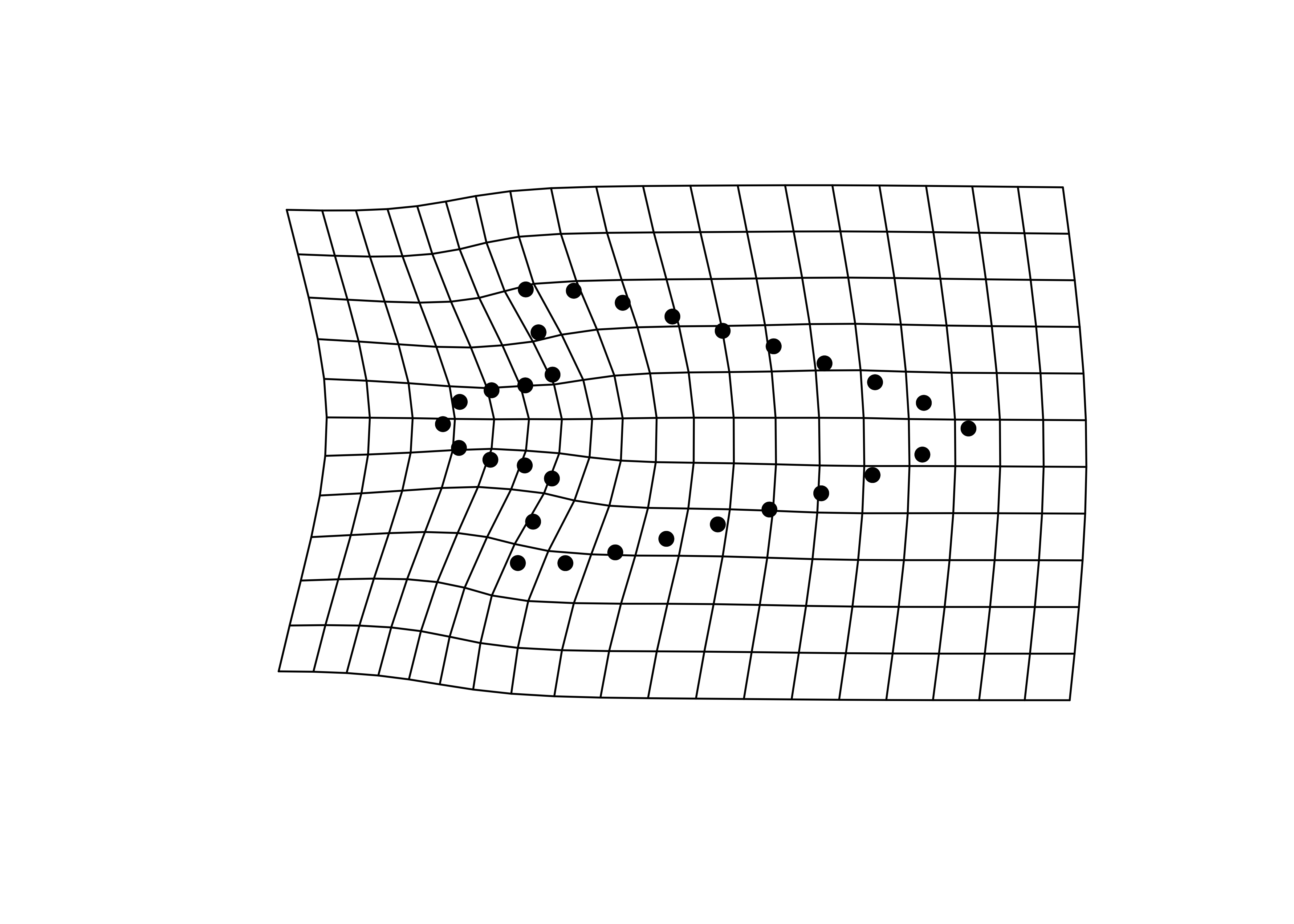
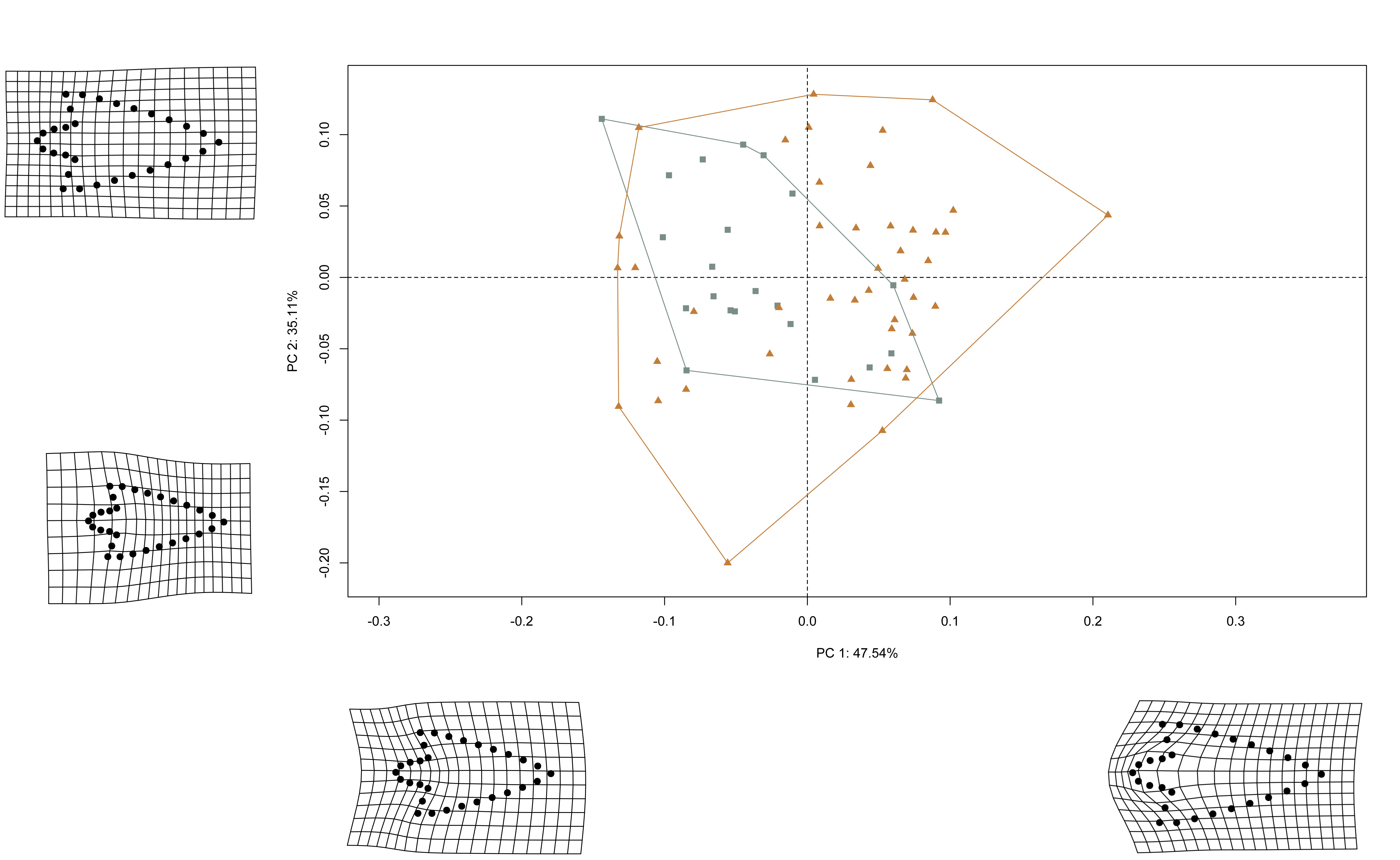
shapes <- readShapes("shapes")
shapesGM <- readland.shapes(shapes, nCurvePts = c(10,3,5,5,3,10))
# read qualitative data
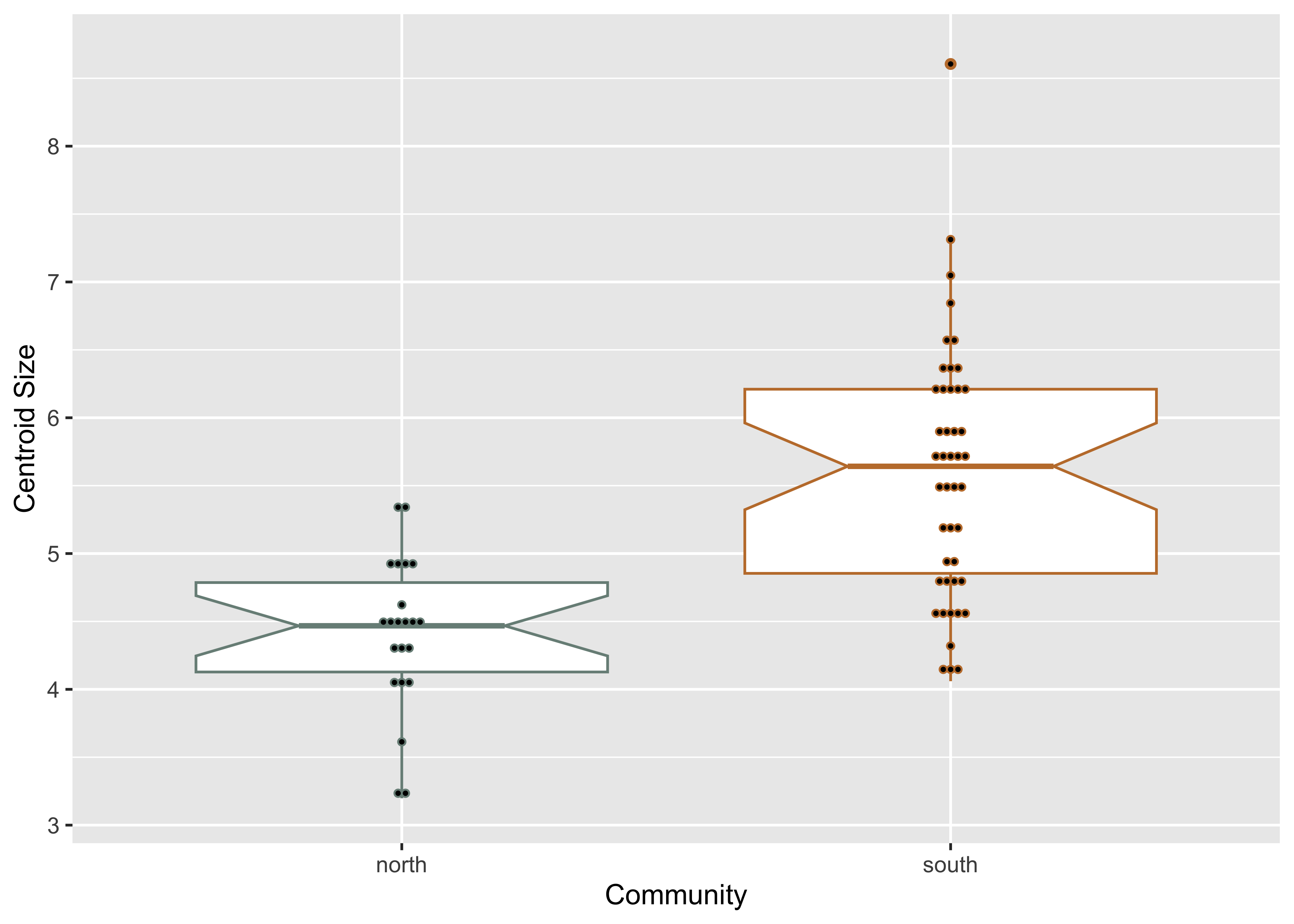
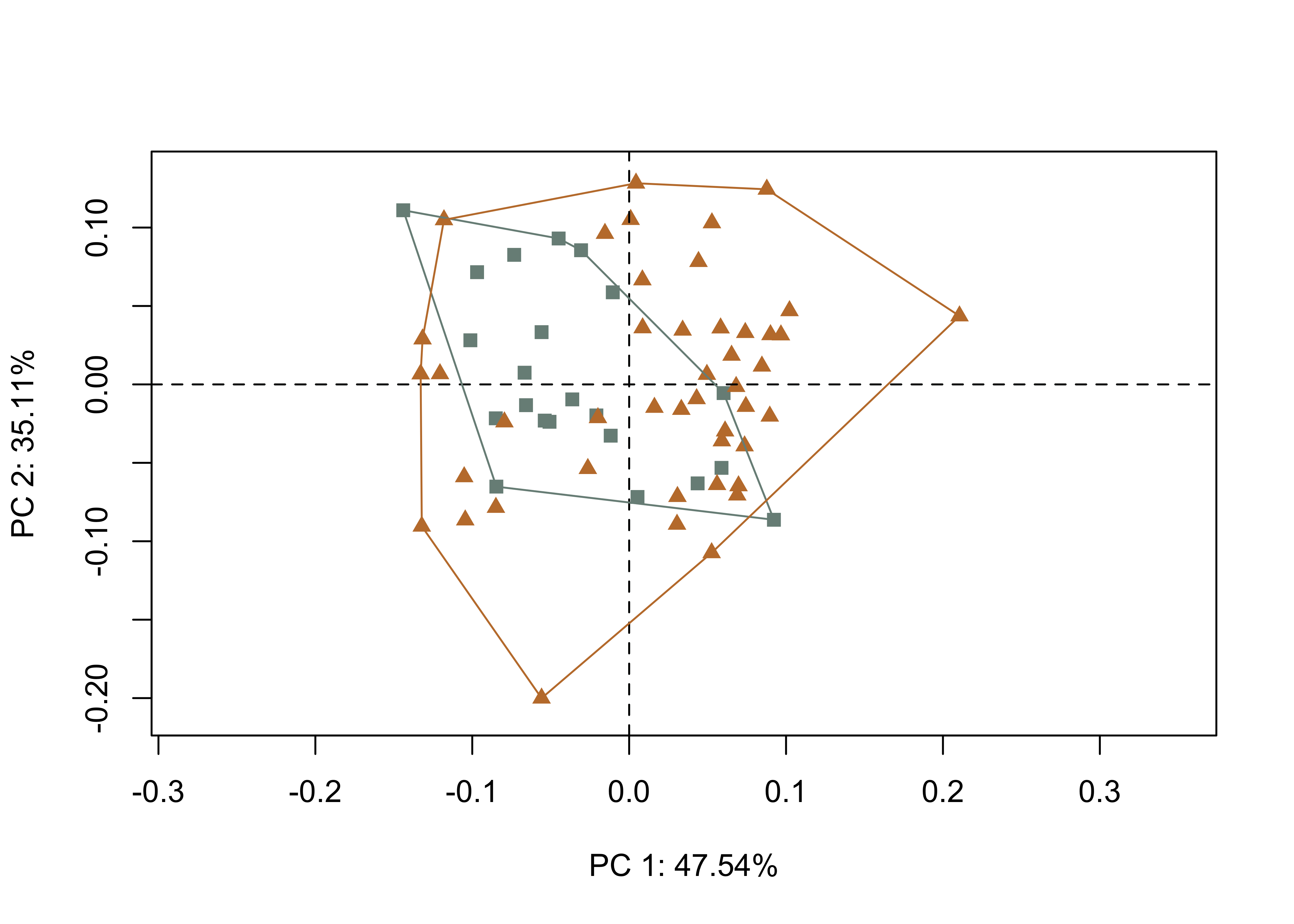
qdata <- read.csv("qdata.csv",
header = TRUE,
row.names = 1)